 NA-MIC Project Weeks
NA-MIC Project Weeks
Back to Projects List
DICOM Segmentation Optimization
Key Investigators
- Steve Pieper (Isomics, Inc)
- Andrey Fedorov (BWH)
- Andras Lasso (Queens)
- Marco Nolden (DKFZ)
- Ralf Floca (DKFZ)
- Hans Meine (MeVis)
- Alireza Sedghi (Radical)
- Erik Ziegler (Yunu)
- Markus Hermann (Idependent)
- Chris Bridge (MGH)
- David Clunie (PixelMed Publishing)
- Sean Doyle (Independent)
Project Description
Discuss our experiences and thoughts on the DICOM SEG standard.
Objective
Compare notes, benchmarks, and experience with interoperability and performance of DICOM SEG instances across platforms. Evaluate the extent to which any observed performance issues are inherent in the format or simply inefficient implementations. Consider proposals to improve the standard to address any inherent issues.
Approach and Plan
- Collate experiences from any investigations and benchmarks to date
- Meet at project week with those on site involving remote participants as possible
- Add notes here about results and plans for any follow up proposals to add representations to the standard
- Discuss if we should consider re-starting/re-thinking the DICOM4QI initiative as a venue to openly promote DICOM interoperability testing and collect feedback from the community.
Progress and Next Steps
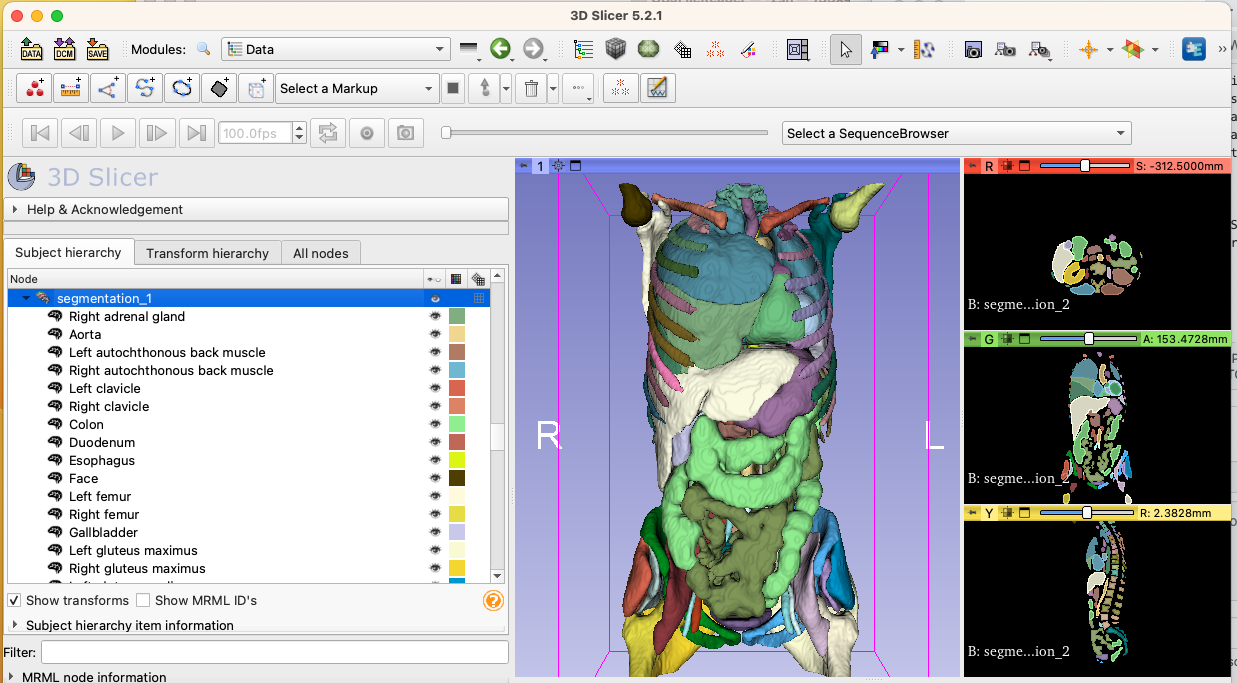
Performed timings with various methods to load segmentations in Slicer
- Quantitative Reporting: 4 minutes
- the underlying issue is due to saving each segment into separate file by dcmqi, this issue is being addressed in https://github.com/QIICR/dcmqi/pull/464
- pydicom-seg: 15 seconds
- seg.nrrd: .6 second
We had several conversations about the importance of DICOM for organizing derived data from quantitative analysis, conversations which underlined the point of defining efficient implementations.
In discussion with machine learning researchers, e.g. developers and users of tools like TotalSegmentator, the number of segments is set increase rapidly, perhaps doubling within months to 200 or more, and with over 1000 segments expected within a year.
Illustrations
Example code to load with pydicom-seg vi Slicer 5.2.1 python console:
try:
import pydicom_seg
except ModuleNotFoundError:
pip_install("pydicom_seg")
import pydicom
import pydicom_seg
import SimpleITK as sitk
dcm = pydicom.dcmread('/Users/pieper/slicer/latest/pydicom-seg/ABD_LYMPH_008_SEG.dcm') # 19 seconds
reader = pydicom_seg.MultiClassReader()
result = reader.read(dcm)
image_data = result.data # directly available
image = result.image # lazy construction
sitk.WriteImage(image, '/tmp/segmentation.nrrd', True)
seg = slicer.util.loadSegmentation('/tmp/segmentation.nrrd')
for segmentID in seg.GetSegmentation().GetSegmentIDs():
segmentIndex = int(segmentID.split("_")[1])
description = result.segment_infos[segmentIndex].SegmentDescription
seg.GetSegmentation().GetSegment(segmentID).SetName(description)
Background and References
The DICOM SEG standard has been around for several years and has been implemented as part of several tools in various languages:
- dcmqi in C++ uses DCMTK and provides support for SEG read/write through the Quantitative Reporting extension to 3D Slicer
- dcmjs supports read/write of SEG in javascript for use in OHIF
- highdicom supports read/write of SEG in python:
- Creation of segmentations from masks numpy arrays (optionally as label map), with metadata copied from source image(s)
- Support for 2D single frame images, image series in patient coordinate system (CT, MR, …), and multiframe tiled images (whole slide images) in slide coordinate sysmte
- Support for images aligned with source images, and in arbitrary space
- Support for “reconstructing” segmentation masks from the stored frames given arbitrary subsets of frames and segments, including conversion to label map
- Note that this branch (under construction) has a much better user guide for SEGs than the current documentation. I.e. this page
- pydicom-seg is using pydicom and SimpleITK, writes single output file. Performance for the TotalSegmentator sample is very good!
- (others - please add to this list)
Known Issues
1. Performance
While interoperability has generally been good, performance of these SEG implementation has in general been orders of magnitude slower than research formats (e.g. nii.gz, nrrd, or seg.nrrd) at supporting segmentation use cases such as using segmentation data for machine learning. For example, this notebook shows that decoding a TotalSegmentator result from DICOM SEG with approximately 100 segments can take several minutes and consume very large amounts of memory for a segmentation that takes less than a second to read from a research format.
Poor performance is due to at least two factors:
- Sub-optimal algorithms/implementations that do not scale.
- The currently released version of highdicom (as of 24th Jan 2023) has an implementation that was never designed to scale. This pull request should make significant progress to address this. Some further improvements should be possible.
- At the pydicom level, iterating through long sequences is slow. This limits the performance of the higher level highdicom because the Per-Frame Functional Groups Sequence can get large in large segmentations. There may be optimisations to make there. See this issue
- Lack of “label map” style encoding in the standard. This is an issue in its own right (see below).
We are interested in how the benefits of DICOM (standardized encoding, rich metadata, coded concepts, etc) can coexist with efficient read-write performance for real-world use cases.
2. Lack of “Label Map” Style Encoding
A DICOM SEG may contain many segments (elsewhere known as “classes” or “labels”). But these segments are each stored in separate frames in the segmentation as multiple binary masks (0 or 1 everywhere). This is in contrast to many other formats that use a “label map” style encoding in which a single array contains many segments using pixel values to represent membership of a segment (i.e. pixel value 1 for segment 1, pixel value 2 for segment 2). Using separate frames does confer two important advantages over the label map approach:
- Segmentations in which the segments overlap each other can be represented
- Fractional segmentations for multiple segments can be represented
However, this also comes at a steep cost for what is arguably the overwhelmingly common use case of non-overlapping non-fractional multi-segment segmentations. Especially in the case of a large number of segments (such as the TotalSegmentator mentioned above), this can lead to a very large number of frames and makes the memory/storage utilization much higher than would be necessary with a “label map” style. When you imagine doing instance segmentation of cells in a whole slide image, this becomes completely untenable.
It has been proposed that this could be solved relatively simply by adding a new Segmentation Type (e.g. “LABELED”) in addition to the existing “BINARY” and “FRACTIONAL”. This is not a formal proposal at this stage.
There is a highdicom draft implementation of what this could look like.
One issue is that currently SEGs images are limited to 8 bits per pixel, which would limit the number of segments representable in “LABELMAP” style to 255. This may not be high enough for some applications (e.g. instance segmentation). A proposal on “label map” encoding should consider whether this limitation should be relaxed.
3. Limited precision for fractional segmentations
Fractional segs are quantized and stored as integers. As mentioned above, the bits allocated is limited to a maximum of 8 currently. This means that fractional segmentations have limited precision and are quantized to 256 values, which is a lower level of precision than users would generally expect.
4. Lack of compression in current implementations
Even if it is encoded in labelmap representations, uncompressed data is inefficient for storing segmentation data. A typical nii.gz or .seg.nrrd file is compressed with gzip and can be 100 or more time smaller than the source data due to redundancy in the segmentation data (large areas of uniform segmentation or repeating patterns that can be more efficiently represented by short codes). DICOM currently offers some options for this like RLE, but as yet they have not be widely supported in currently used open source tools.
5. Some interoperability concerns
There are repeated reports of interoperability issues between segmentations created with highdicom and viewed in OHIF. See this issue.
6. Expanding dimension organization methods
Multiple users of highdicom have been asking for support for 2D+T files. This is possible but not straightforward due to the need to create a dimension organization methodology that includes time as a dimension. Due to time limitations this has not been a priority for highdicom but remains an open issue. See
- Higidicom issue 200
- Higidicom issue 174
- Some related discussion here
A broader issue is whether these would be understood by viewing software unless the dimension organization method is standardized to some extent.